Research
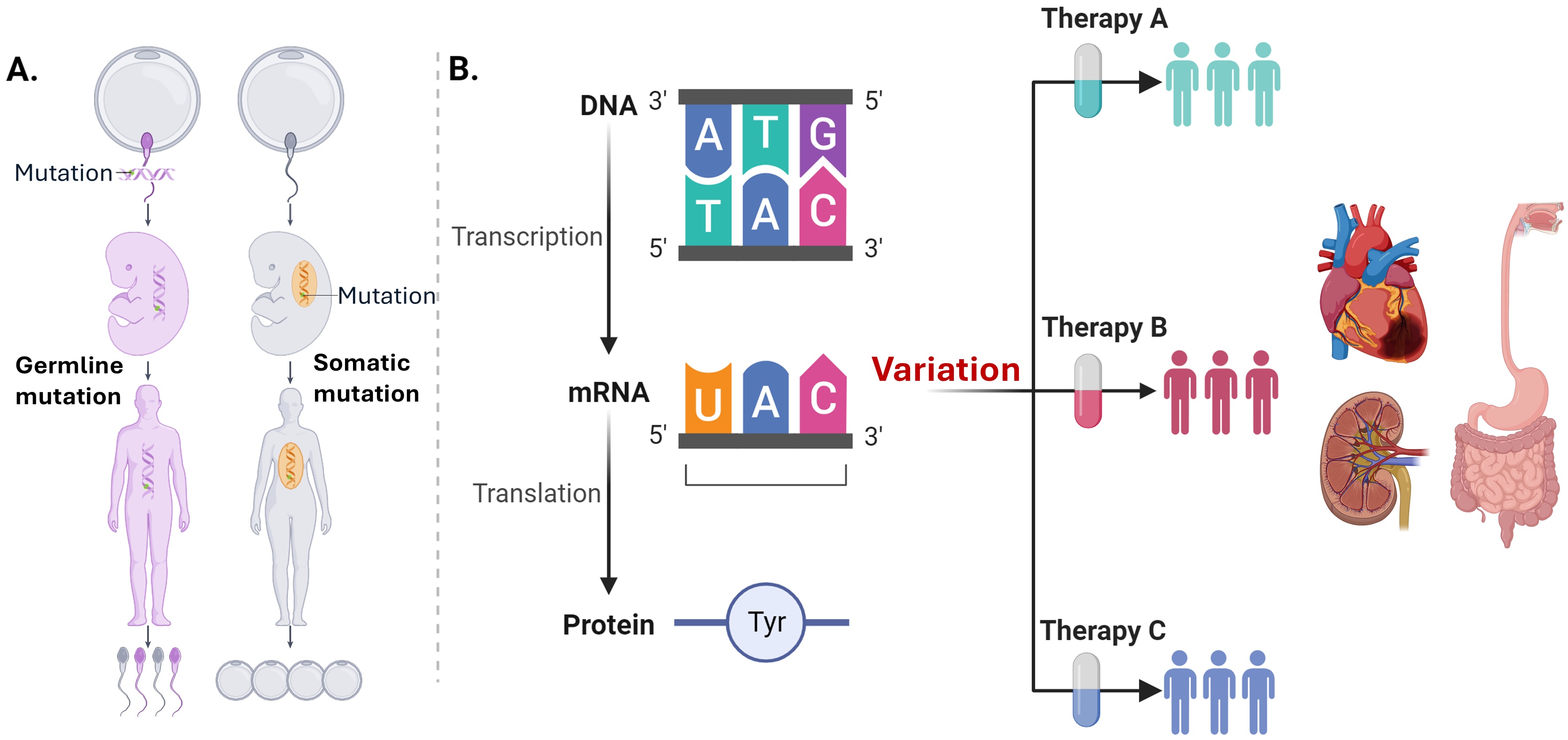
Genetic variations underpin human phenotypic differences, including susceptibility to age-related diseases. These variations occur along two key axes: (A) The first axis involves DNA, differentiating between germline mutations, which we inherit, and somatic mutations, which are acquired as we age and localized to specific cells; (B) The second axis follows the central dogma, where changes in DNA affect RNA and subsequently lead to variations in protein levels. Together, these omics form biological processes, and the variability at each level contributes to an individual’s unique health profile, laying the foundation for personalized medicine, and this is what our research builds on.
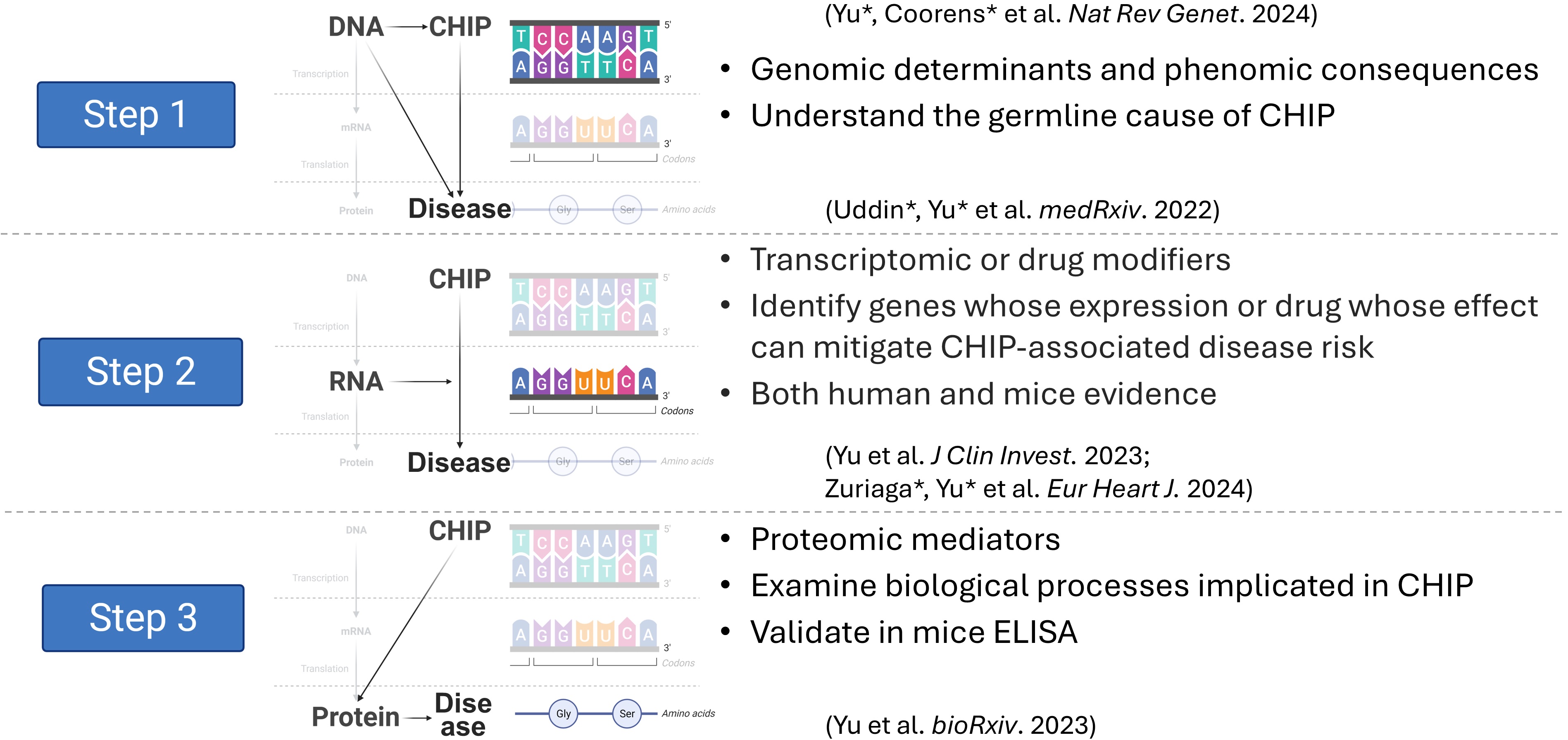
Age-related clonal hematopoiesis and its somatic-germline interactions, with implications for managing CH-associated diseases
Age-related clonal hematopoiesis (CH) arises from somatic mutations that drive clonal expansions within the hematopoietic system. Mutations frequently occur in genes such as DNMT3A, TET2, ASXL1, and JAK2. While CH is a precursor to myeloid malignancies, numerous studies in both human and model systems link CH to a broader spectrum of conditions, including cardiovascular and other age-related diseases, though the underlying mechanisms remain largely unclear. The mutated genes implicated in CH exhibit distinct multi-omic profiles, necessitating a comprehensive integrative approach. To uncover the mechanistic connections between CH and these diverse diseases, our research traces the central dogma (DNA, RNA, protein) to unravel how somatic mutations contribute to pathogenesis.
Relevant works include: Uddin*, Yu* et al. 2022, Yu et al. 2023, Zuriaga*, Yu* et al. 2024, Yu et al. 2023, and Yu*, Coorens* et al. 2024.
Omics-based instruments for testing causal hypothesis in studying diseases
Germline genetics are intrinsic causes for postnatal diseases, forming the basis of Mendelian randomization (MR) for causal inference. Our research involved leveraging or refining MR methods for more robust insights into cardiovascular and other age-related diseases. We also build predicted RNA expression level use DNA and integrate the resulted instruments for examining somatic-germline interplay.
Relevant works include: Yu et al. 2022, Yu et al. 2022, Yu et al. 2020, Jin et al. 2024, and Yu et al. 2023.
Disease risk prediction using static (germline genetic) and dynamic (time-varying fators) data
Our research adapts or develops tools for predicting individual disease risk by leveraging both static (germline genetic) and dynamic (time-varying lifestyle and clinical) data. Germline genetics account for a substantial portion of variability in human traits, and our work leverage that to create polygenric risk score (PRS) for the prediction of disease of interest. For example, our PRS for kidney function remained the best performed one in European ancestry so far. Beyond static genetic factors, dynamic variables such as changes in lifestyle and health history also significantly influence disease outcomes. We improved disese risk prediction by adapting machine learning models to include time-varying covariates and enhanced the interpretability of ‘black box’ predictions.
Relevant works include: Yu et al. 2021, Yu et al. 2022, Steinbrenner et al. 2023, and Zhang et al. 2023.
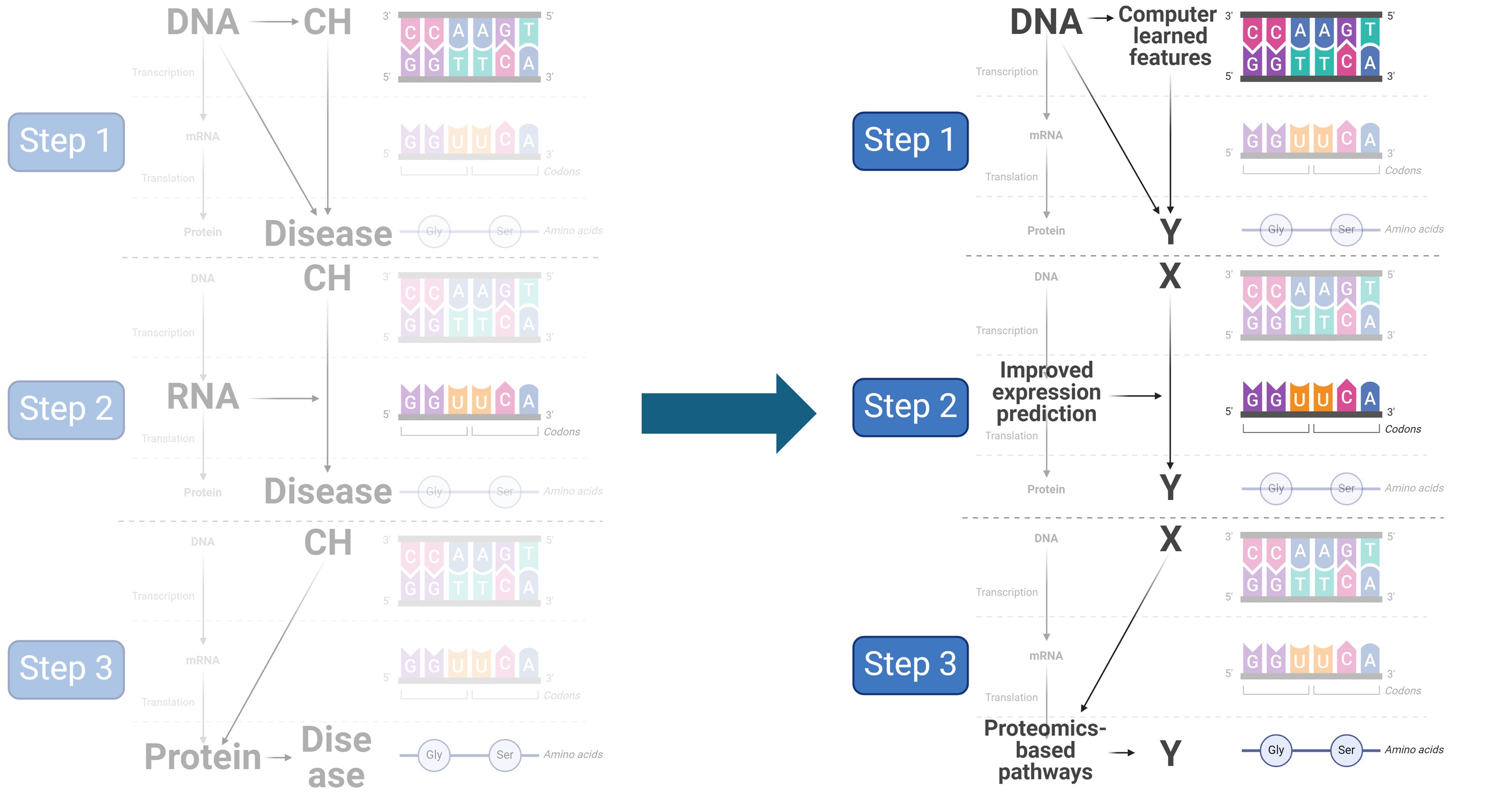
Machine learning + multi-omics
Our ongoing work integrate machine learning with multi-omics, also spans DNA, RNA, and protein levels. At DNA level, we examine the genetic basis of computer-learned features from medical imaging. At RNA level, we are fine-tuning foundational models for improving their understanding of DNA loci in modifying gene expression variability. At protein level, we are developing methods for building protein-based pathway for improving translational insight.
Relevant work include: Kamineni…Yu*, Natarajan* 2024